Cancer-specific Alternative Splicing Events
Novel diagnostic and prognostic biomarkers for different cancer types
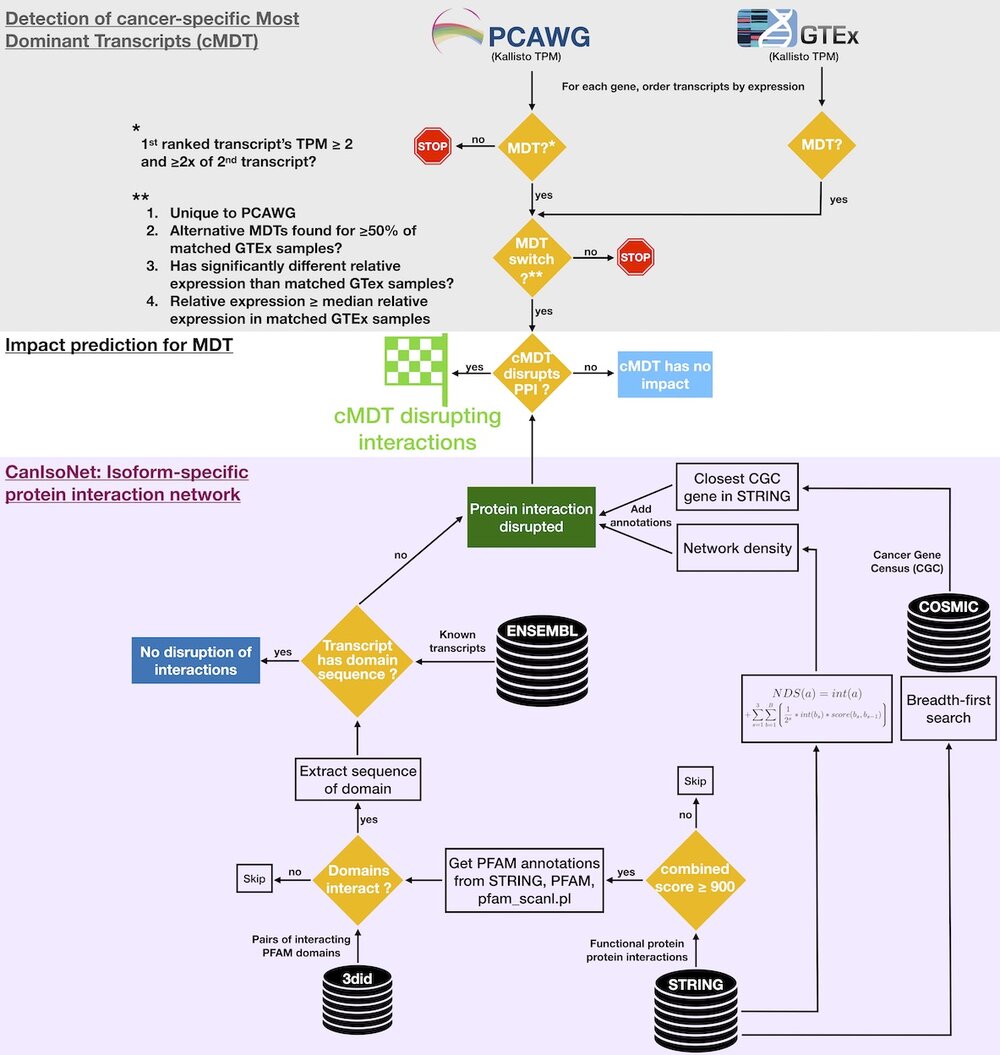
Alternative RNA splicing is a regulatory cellular mechanism to create multiple mRNA molecules from the same gene and is often disrupted in diseases. The Data Science in Life Science Team headed by Dr. Abdullah Kahraman could show in the international Pan-Cancer Analysis of Whole Genomes (PCAWG) study that such disruptions in alternative splicing patterns are widespread in cancer. Based on these results, they have started to search for novel diagnostic and prognostic biomarkers for superior diagnostic, treatment, and medication for cancer patients.
Tools and Databases
To aid the search the team behind Dr Kahraman is developing different tools and databases related to identifying pathogenic alternative splicing events. These databases include CanIsoNet, a database visualizing protein-protein interaction losses of disease-specific isoforms, IsoAligner, a web service to map amino acid positions across protein isoforms, and a novel isoform-specific drug-interaction database.
The first publications of their tools and work are available here:
Hanimann, J., Moch, H., Zoche, M. and Kahraman, A. (2022). IsoAligner: dynamic mapping of amino acid positions across protein isoforms. F1000Research 2022, 11:382
Karakulak, T., Szklarczyk, D., Moch, H., von Mering, C., Kahraman, A. (2021). CanIsoNet: A Database to Study the Functional Impact of Isoform Switching Events in Cancer. bioRxiv
Kahraman, A., Karakulak, T., Szklarczyk, D., Mering, von, C. (2020). Pathogenic impact of transcript isoform switching in 1,209 cancer samples covering 27 cancer types using an isoform-specific interaction network. Sci. Rep. 10, 1–15.
Comments
No comment posted about Cancer-specific Alternative Splicing Events